Running CIVET on ChRIS at FNNDSC
CIVET is a T1 MRI processing pipeline (similar to FreeSurfer) developed at the Montréal Neurological Institute (MNI). This tutorial is a beginner-level walk-through of how to run CIVET on ChRIS at the FNNDSC.
1. Log in to ChRIS
Go to http://chris-next.tch.harvard.edu:2222/login and log in using your FNNDSC account username and password.
2. Create a feed
Data are imported into ChRIS in feeds.
- Click "Create Feed" in the top-right corner.
- Choose the "Generate Data" option.
- Select the
pl-neurofiles-pullplugin, which pulls data from/neuro/labs/grantlab/researchinto ChRIS. - Specify which path to pull as the
--pathparameter. - Keep clicking "Next" until the feed is created.
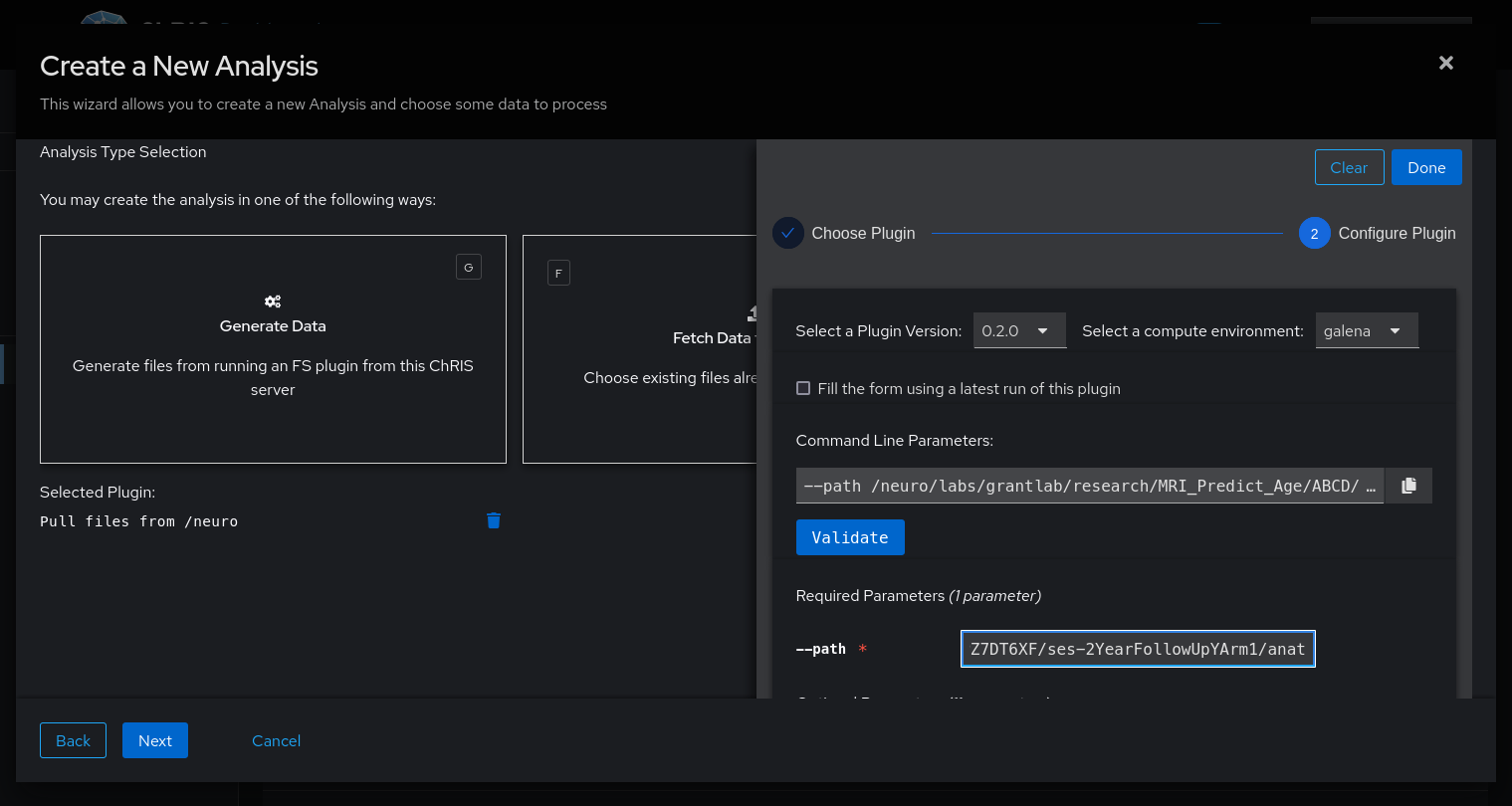
The path you choose must be a directory containing exactly one NIFTI file called *_T1w.nii.
Any unrelated files (such as *.json, *._T2w.nii) will also be uploaded to ChRIS but ignored by the pipeline.
3. Run the pipeline
Click into the feed you just created, and right-click the only circle in the top-left quadrant. Select "Add a pipeline."
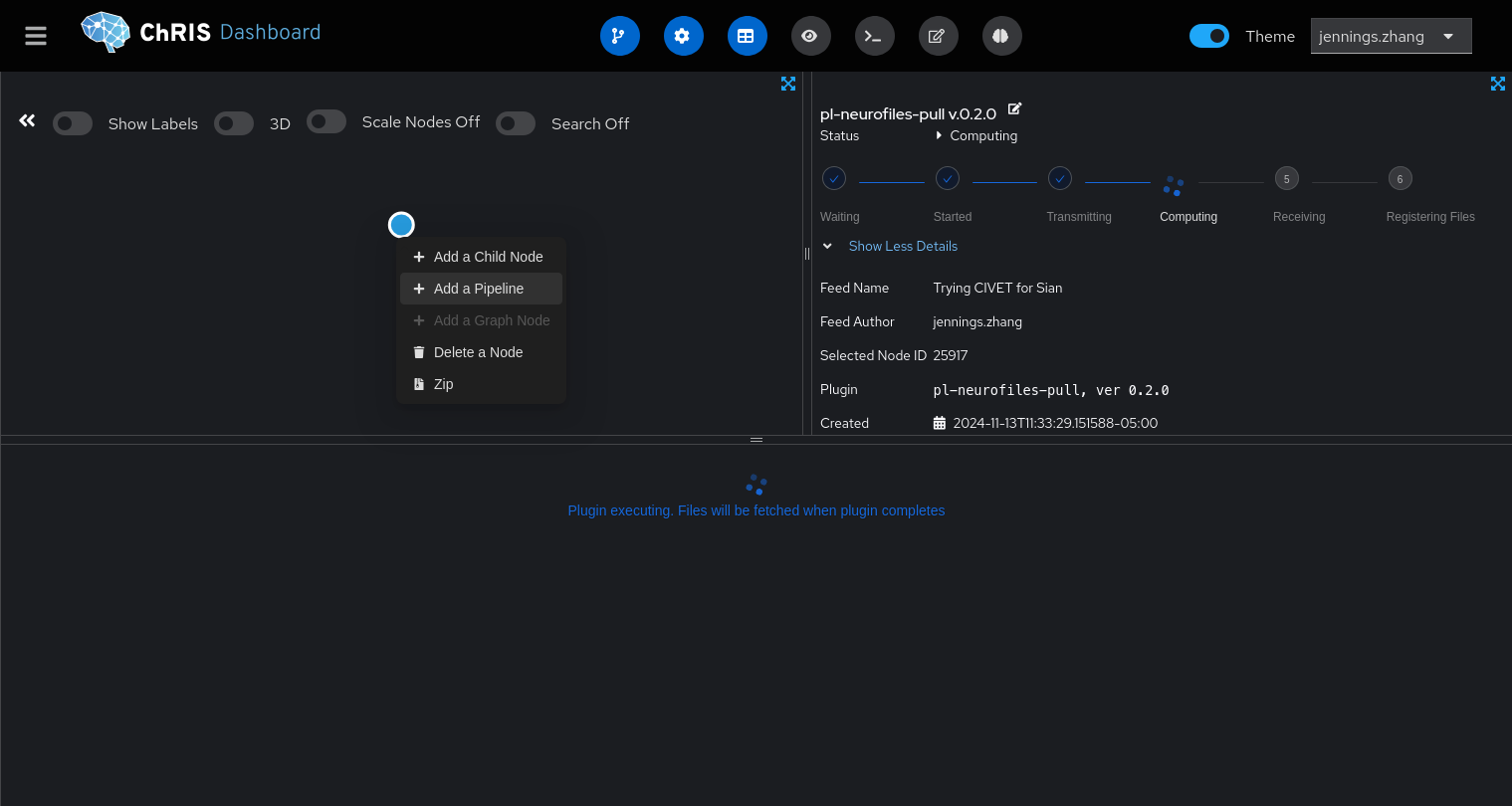
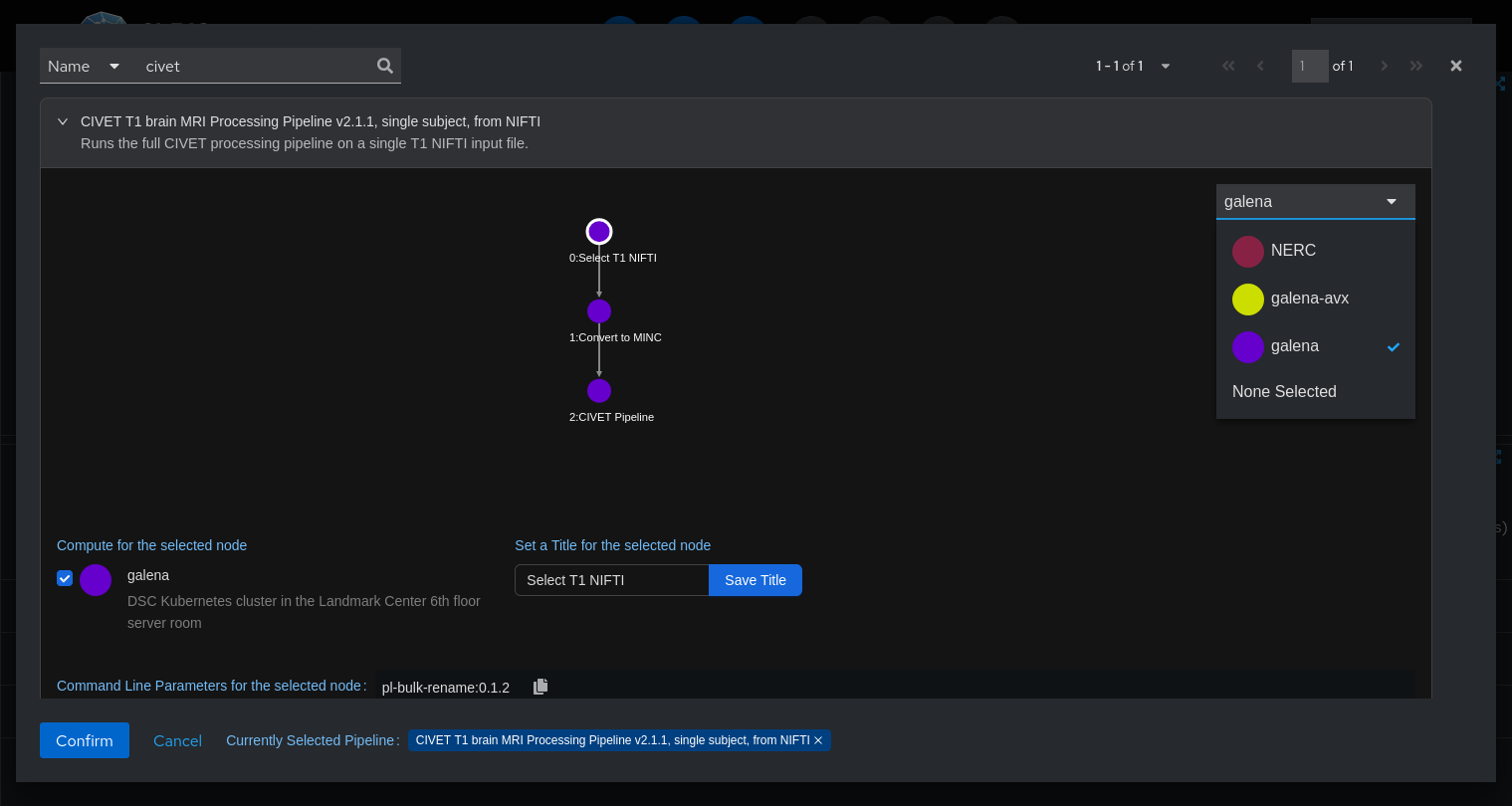
Search for the "CIVET" pipeline and select it.
Carefully review the pipeline's options before you run it. Make sure that each step of the pipeline is configured to run on "galena," our private compute cluster. You may also choose "NERC," a public cloud, as long as your data does not contain PHI.
Once the pipeline is running, you can see each of its steps and their statuses. You can see that the pipeline selects the *_T1w.nii file, renames it to *_t1.nii, then converts it to MINC.
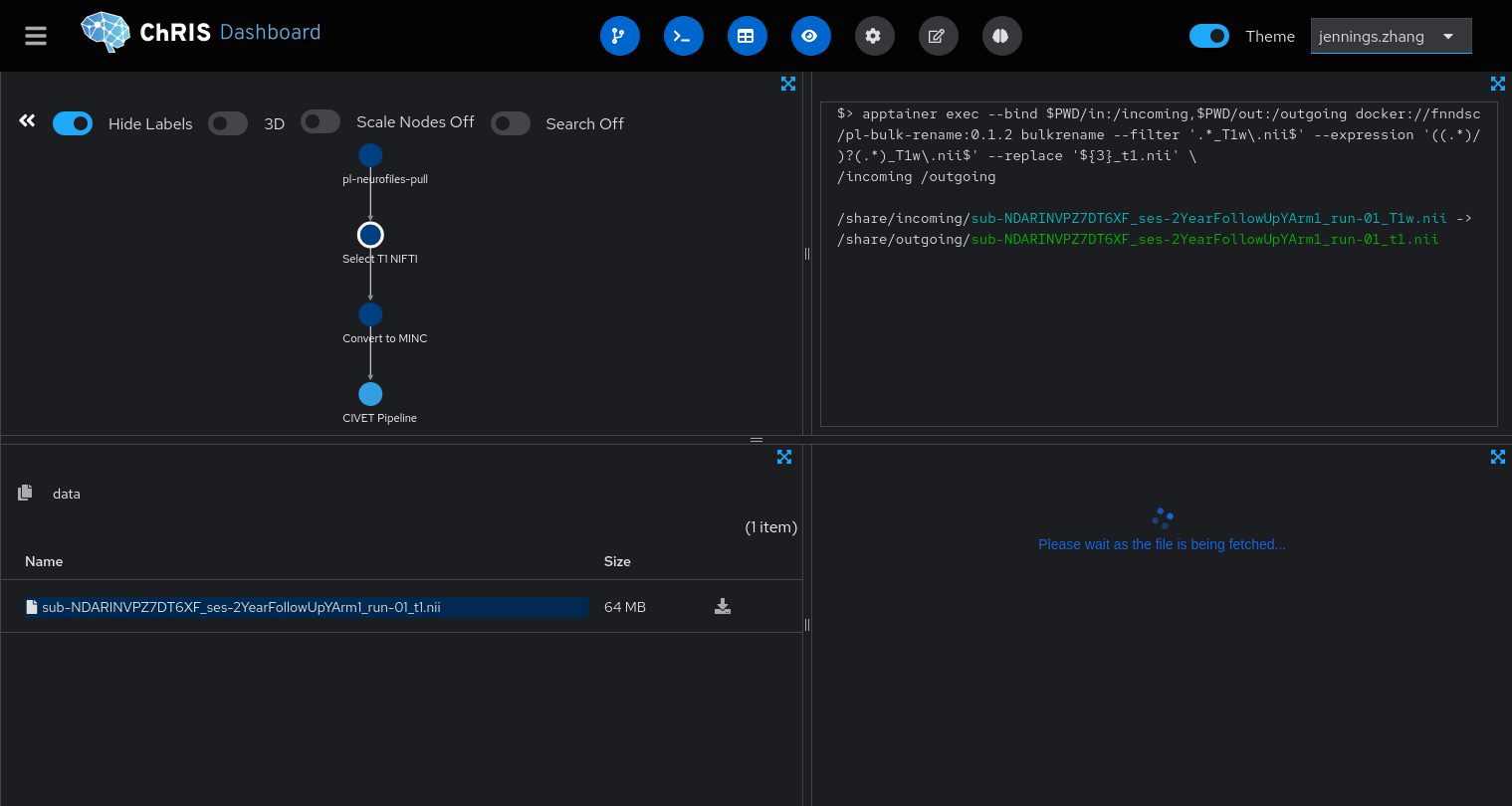
4. Wait a couple hours
CIVET takes 4-8 hours to run to completion.
Next Steps
You now know how to run CIVET using the ChRIS user interface. Next steps might include visualization and downstream analyses of CIVET outputs, or scaling up to run this pipeline on multiple subjects.